Study sheds new light on multi-parent populations’ potential for crop breeding
A recent study by an international team of scientists gives a new insight into how multi-parent populations (MPPs) can be better used for crop research, development and breeding.
Published in Heredity, an official journal of the Genetics Society, the study proposes an ideal package of genotypic, phenotypic and germplasm resources that MPPs should feature to realize their full potential.
Based on the review of previous research on various crops, including pigeon pea, chickpea, faba bean, durum wheat, barley and rice, the study shows that crop populations obtained from multi-parent crosses enable a genetic analysis of complex traits in different crops and facilitate modern as well as conventional plant breeding.
By mixing and recombining the genomes of multiple founders (parents used in crop population development), MPPs combine many commonly sought beneficial properties of genetic mapping populations. For example, they have high power and resolution for mapping multiple quantitative trait loci (QTLs), high genetic diversity and minimal population structure. Over the past few years, several MPPs have been constructed in different crop species, and their inbred germplasm and associated phenotypic and genotypic data serve as powerful resources for breeders.
Moreover, the utility of MPPs has grown from just being a tool for mapping QTLs to a means of providing germplasm for crop breeding programs. The approach is cost-effective and makes it possible to gather valuable genomic information for large crop populations.
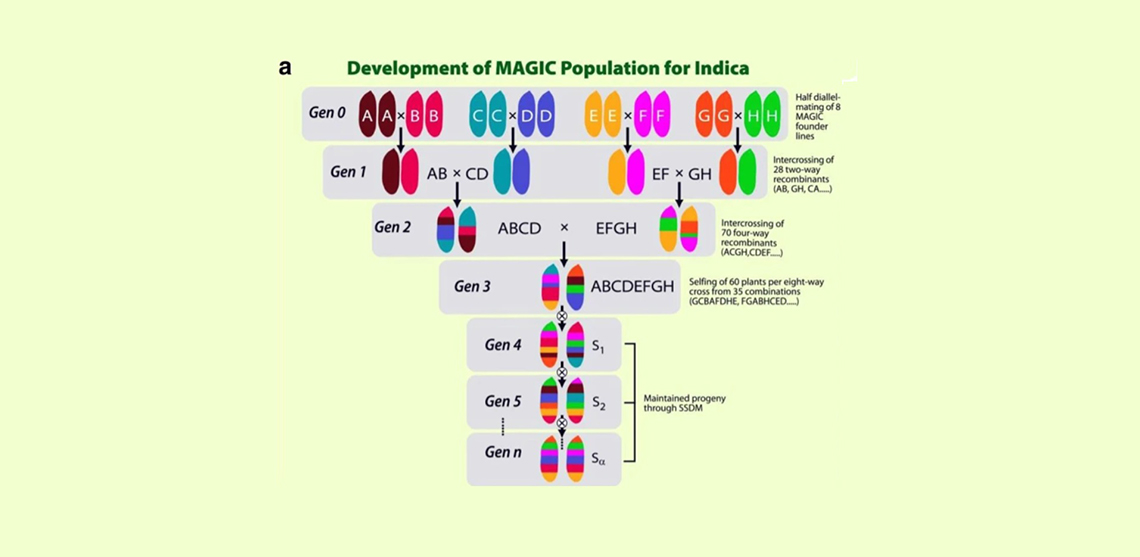
Currently, MPPs in plants have two most popular designs - nested association mapping (NAM) and multi-parent advanced generation inter-cross (MAGIC) populations. Compared to NAM, the MAGIC allows the high-resolution mapping of quantitative traits. MAGIC populations typically descend from 4, 8 or 16 parents, consistent with a simple funnel breeding design. Due to their complex pedigree structure, MAGIC populations offer unique opportunities in crops and have great potential for improving breeding populations and dissecting genomic structure.
Dr. Rakesh Kumar Singh, Program Leader on Crop Diversification and Genetics at ICBA and a co-author of the study, said: “Resulting from crosses between two inbred lines, the bi-parental populations (BPPs) have been mostly a standard design for genetic mapping in crops. However, they have two main disadvantages: the lack of mapping precision, which stems from limited effective recombination (a single event of crossing over) occurring during population development, and low genetic diversity, which is due to the genetic bottleneck caused by choice of two founders. This may limit the number of QTLs captured as no more than two alleles (a variant form of a gene) segregate at any loci. Therefore, MPPs, the second-generation mapping populations, are developed by inter-crossing multiple parents.”
The study presents a comprehensive account of MPPs and their useful applications in crop breeding. It also suggests thinking of MPPs as a ‘package’, ideally comprised of germplasm of the recombinant inbred lines (RILs) and their founders, free of intellectual property constraints.
The study also advocates for a publicly accessible database of agronomically important phenotypic data for the founders and the descendent RILs with a system for adding further phenotypic data. To harness the power of MPPs as a genetic toolbox to facilitate crop improvement, there is a need for the continued enrichment of MPPs with open-access phenotypic and genotypic resources.
The study is intended to help researchers fast-track breeding efforts and introduce crop varieties with a desirable combination of multiple traits such as tolerance to biotic (pests and diseases) and abiotic (drought and salinity) stresses.

